Rapid manual nucleic acid isolation using the spin filter method
Nucleic acid isolation with our XXprep Kits works according to the spin filter method. This technique is the extraction method of choice for small sample volumes and a regular or occasional throughput of 1-20 isolations (low throughput). With an extraction time of approx. 20 – 40 min per purification and – depending on the nucleic acid type – a yield of up to 100 µg of highly pure DNA/RNA, the “Miniprep” is the standard method in most molecular biology laboratories (academic research, molecular diagnostics, genetic engineering).
What is important when choosing the right kit?
For the users, factors such as time, simplicity of the protocol, availability of all the required equipment (in this case only a tabletop centrifuge), and the purity of the isolated nucleic acids play a role. Of course, the kit has to fit the sample! It is a big difference whether the aim is isolation of plasmid DNA from bacteria or RNA from sewage. The chemistry and the protocol have to be perfectly matched to each other and may vary considerably with the type of nucleic acid and sample.
Our XXprep kits currently cover the following isolations:
- Genomic DNA from bacteria, tissue, rodent tails, eukaryotic cells, whole blood
- Plasmid-DNA/ Cosmid-DNA from bacteria
- Total RNA from bacteria, eukaryotic cells and tissue
- DNA fragments from agarose gels, PCR reactions and other enzymatic approaches
All XXprep kits achieve very high purity for the isolated nucleic acids. Typical A260/A280 values are 1.8-1.9 for DNA and 2.0 for RNA.
How does it work?
Nucleic acid isolation with the XXprep kits is based on the spin filter method. Looking at the different types of kits, there are major differences regarding lysis and pre-treatment of the lysate. However, the process on the column is by and large identical:
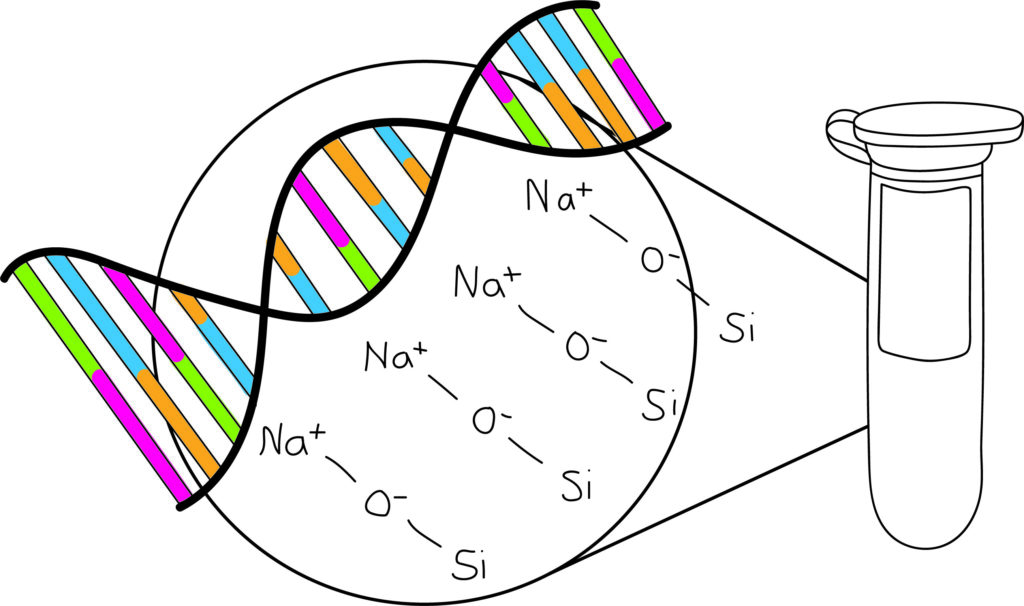
- Binding of the nucleic acid to the solid phase takes place with a combination of chaotropic and non-chaotropic salts of low ionic strength. Na+ ions mediate the bonding between the negative surface of the silica matrix (SiO–) and the phosphate groups of the nucleic acids.
- Washing with low-salt buffers flushes impurities from the column.
- Elution takes place under salt-free conditions. The eluted nucleic acids are thus perfectly suited for any kind of downstream applications, e.g. cloning (restriction digestion), PCR, sequencing, transfection and in vitro transcription.
Advantages of our XXPrep Kits
- Easy to perform according to spin filter method
- High yields
- Very high purity
- Reduced use of plastics
Overview: The most important details on the individual XXprep Kits
Code 4427, Plasmid Mini Kit for rapid isolation of plasmid and cosmid DNA from bacterial cultures of up to 15 ml.
- Reduced use of plastics
- Novel filter membrane with increased binding capacity enables yields of up to 80 μg of highly pure plasmid DNA (high-copy and low-copy plasmids)
- High yields and excellent purity
The plasmid DNA is extracted from the bacterial cells by an alkaline lysis procedure and (following precipitation of genomic DNA and proteins) bound to the silica membrane of a spin column. The plasmid DNA is released from impurities by various washing steps and then eluted with 50 – 100 µl buffer.
Technical data
Starting material
- Bacterial culture of 0.5 – 15 ml
Binding capacity of the column
- 80 µg DNA
Time required
- 15 min
Yield
- 6 – 20 µg (high-copy plasmids) from 2 ml bacterial culture
- 40 – 80 µg (high-copy plasmid) from 15 ml bacterial culture
Code 2710, Kit for the rapid and efficient isolation of genomic DNA from various amounts and types of starting material, such as tissue samples, mouse or rat tails, bacterial and eucaryotic cell pellets, and whole blood.
- Variety of sample types
- Reduced use of plastics
- Protocols for 200 μl and 400 μl whole blood
- High yields and excellent purity
The extraction procedure combines the lysis of various starting materials with the subsequent purification of the DNA via a silica column.
Note: The standard protocol isolates RNA as well as DNA. Users who require RNA-free DNA can optionally perform an RNase digestion before applying the sample to the column.
Technical data
Starting material
- Tissue samples up to 20 mg
- Whole blood (fresh or frozen; stabilized with EDTA or citrate) up to 400 µl
- Rodent tail pieces 0.2 – 0.8 cm
- Cell cultures up to 5 x 106 cells
- Bacterial cultures up to 1010 cells
Binding capacity of the column
- 100 µg DNA
Time required
- approx. 10 min after lysis
Yield
- Depending on sample material; can be more than 100 µg DNA per column
Purity
- A260/A289: 1.8 – 2.0
- A260/A230: 1.8 – 2.3
Code 5482, Kit for the rapid purification of PCR products in a simple two-step procedure and/or for the extraction and recovery of DNA fragments from TAE or TBE agarose gels.
- Combi Kit
- PCR purification in a two-step process (without unnecessary washing)
- Reduced use of plastics
- flexible elution volumes (10 – 50 μl)
- suitable for fragment lengths from 100 bp to 30 kb
- High yields and excellent purity
Actually, it is the same as with genomic DNA, except that a special binding buffer is used here (optimized for smaller pieces of DNA). In addition, the surface functionalization of the silica matrix is different.
Technical data
Starting material
- PCR and sequencing reaction mixtures up to 50 µl
- TAE or TBE agarose gels up to 300 mg
- Fragment lengths of 100 bp – 30 kb
Binding capacity of the column
- 20 µg DNA
Time required
- PCR and sequencing purification approx. 3 min (2-step procedure)
- Gel extraction approx. 20 min
Recovery rates
(Depending on fragment length)
- PCR purification 60 – 95 %
- Sequencing purification min. 75 % 75 %
- Gel extraction 60 – 90 %
Code 6780,Kit for the rapid and efficient isolation oftotal cellular RNA from eucaryotic cells, bacteria and tissue samples.
- Variety of sample types
- Selective removal of genomic DNA without DNase digestion
- No toxic and foul-smelling ß-mercaptoethanol required
- Reduced use of plastics
- High yields and excellent purity
A specially optimized lysis buffer system guarantees the isolation of intact RNA with permanent deactivation of endogenous and exogenous RNases. By means of a first column, the genomic DNA is removed, which makes DNase I digestion no longer necessary. The RNA is bound to a second silica column, washed and finally eluted in 30 – 80 μl of RNase-free water.
Attention: The kit is not suitable for viral RNA! For the lysis of viruses, a different lysis buffer system is needed.
Technical data
Starting material
- Tissue samples (biopsies) up to 20 mg
- Cell cultures up to 5 x 106 cells
- Bakterienkulturen bis zu 109 Zellen
Binding capacity of the column
- 100 µg RNA
Time required
- 15 – 40 min
Yield
- Depending on sample material; can be up to 100 µg DNA per column
Purity
- A260/A289: 1.8 – 2.1

